The following method works in TopSpin 2. It works with some changes in TopSpin 1. It hasn’t been tried in xwinnmr, but it should work.
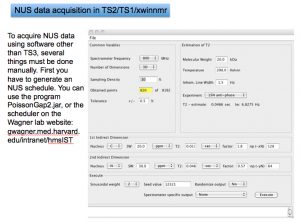
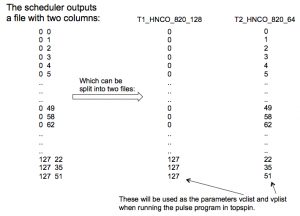
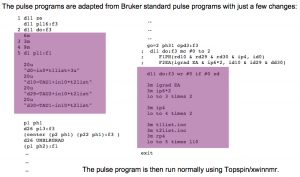
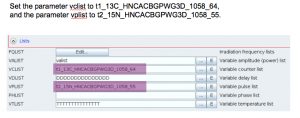
Example: an HNCACB with 64 (uniform) complex points in F1 (13C) and 55 (uniform) complex points in F2 (15N). 30% sampling –> 1058 hypercomplex NUS points in the sampling schedule. The 2 columns of the sched file are split into 2 separate files:
- t1_13C_HNCACBGPWG3D_1058_64 –> /opt/topspin/exp/stan/nmr/lists/vc
- t2_15N_HNCACBGPWG3D_1058_55 –> /opt/topspin/exp/stan/nmr/lists/vp
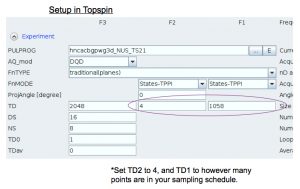
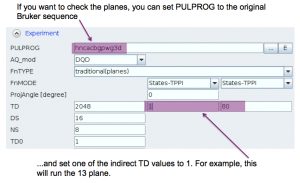
Then run normally with zg.