*Note: this suite of experiments works best on TS3 and higher.
A very useful suite of experiments was published and made available by the Veglia and Kalodimos groups:

There are 24 separate experiments, most of which correlate two or more of the following: H/C-methyl, H/N-amide, H/C-aromatic. There are four 2Ds and twenty 3D NOESYs that correlate various combination of these nuclei.
*The setup of these experiments is completely automated. No user input is needed except for routine things like NS, SW, etc.
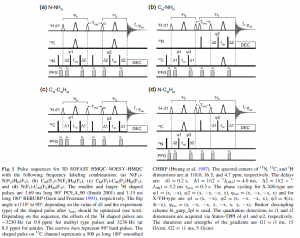
To set these experiments up, start from any dataset with a calibrated proton pulse. Just type “sofast.py” in the topspin command line. It will bring up a window asking you to confirm the proton 90 degree pulse length and power, and the value of o1p. Then you have to select an experiment number. Choose one from this list:
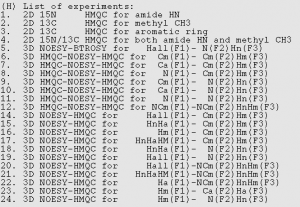
…and that’s it! The only other things you need to set are sample-specific things like sweep width, number of scans and increments, etc. Don’t change any of the pulse settings.