by Robert Peterson – 09/2020 – UCLA
The fluorine nucleus (19F) has some unique advantages as an NMR probe for biological systems.
- 19F is a spin-1/2 nucleus
- Its natural abundance is 100%
- It has the second-highest gyromagnetic ratio, after 1H. It’s intrinsic signal-to-noise is 83% as high as proton.
- It has a very broad chemical shift range (>400ppm), which makes it very sensitive to subtle changes in chemical environment
- Fluorine is almost completely absent from biological systems, but can relatively easily be introduced as a probe.
- Fluorine substitution usually has a minimal effect on structure and function.
And: our new probe on the 800 MHz NMR has a 19F channel.
Fluorine NMR is a powerful tool to investigate dimerization, ligand binding, dynamics, protein folding, aggregation and misfolding, etc in a relatively straightforward way. That is: without getting caught up in the weeds of complicated and time-consuming multidimensional experiments. Most of the fluorine NMR studies can be done with simple one-dimensional experiments.
Example: study of α-synuclein fibrillation done with 19F NMR
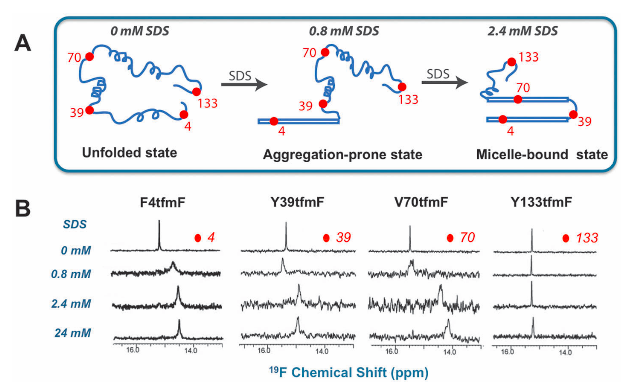
Fluorine labeling of proteins
There are many methods to fluorinate proteins. The most common amino acids that are labeled with fluorine are the aromatic amino acids. These are a good choice because most protein sequences contain few aromatic residues, so there usually aren’t very many to assign. Also, aromatic amino acids tend to be overrepresented in structurally interesting regions of proteins, such as active sites and dimer interfaces. Another common way to incorporate fluorine into proteins is to conjugate fluorine-containing small molecules to protein side chains. Cysteine labeling has been the most common of these methods.
These fluorinated amino acids, and many others, are available commercially:
- 5-F-L-Tryptophan
- 6-F-L-Tryptophan
- 3-F-L-Phenylalanine
- 2-(Trifluoromethyl)-L-Phenylalanine
- 3-(Trifluoromethyl)-L-Phenylalanine
- 4-(Trifluoromethyl)-L-Phenylalanine
- 4-F-L-Phenylalanine
- 3-F-L-Tyrosine
This document contains three laboratory protocols representing, hopefully, the three simplest and most useful methods to fluorinate proteins.
The first protocol describes a method for labeling proteins with fluorinated versions of any of the three aromatic amino acids. In it, glyphosate is used to inhibit the cell’s production of the aromatic amino acids. The three aromatic amino acids (with one or more fluorinated) are also added prior to induction. This method is the most efficient way to fluorinate phenylalanine or tyrosine, and can also be used to fluorinate tryptophan.
The second protocol describes the most efficient method for labeling proteins with fluorinated tryptophan. In it, 5-fluoroindole is added prior to induction. No inhibitor or other amino acids are needed. According to the authors, it represents a 15-fold decrease in cost for fluorinating tryptophans relative to the first method.
The third protocol describes a method for conjugating a fluorine-containing small molecule onto cysteine residues of a previously purified protein.
Protocol #1: expression protocol for protein labeled with 19F-labeled aromatics
By Robert Peterson, Yuan Yang, and Mark Arbing – 08/2020 – UCLA
*This protocol was adapted from the literature and has not been tried at UCLA
Overview: This method is used for fluorine labeling one (aromatic) amino acid type in a protein. E. coli BL21 cells harboring an appropriate expression plasmid are grown in minimal media. Glyphosate, which inhibits the shikimate pathway and shuts down synthesis of the aromatic amino acids in the cell is added just prior to induction by IPTG. Tryptophan, phenylalanine, and tyrosine are also added, and a fluorine labeled version of whichever one is desired to be fluorinated is used. For example, if 3-fluoro-tyrosine is desired, then 3-F Tyr would be added rather than unlabeled tyrosine. The protein can be purified normally.
Detailed Protocol, using 3-F-tyrosine as an example:
Day 0: Perform a plasmid transformation into E. coli BL21-Gold (DE3) competent cells and plate on LB-Agar supplemented with antibiotic. Incubate the plate at 37 °C for 16-20 hrs. Prepare 1 L of minimal media in 2.8 L baffled culture flasks by diluting 10X M9 salts (refer to steps 1 and 2 in the minimal media recipe below) with deionized water, add 1 g of NH4Cl, autoclave with liquid cycle and store at room temperature. If you wish to generate protein with both 15N and 5-FW label, use 1g of U-15NH4Cl instead of regular NH4Cl.
Day1: Inoculate a 5 mL starter culture at 9am in LB-Antibiotics by picking up a single colony from the transformation plate or by scraping a chunk of glycerol stock. Incubate starter culture by shaking at 205 rpm in a 37 °C shaker. The duration of starter culture shaking should be >3hrs. You can also shake the starter culture overnight (~6-12 hrs).
Add the rest of reagents to the prepared 1 L minimal media (step 3 in the minimal media recipe below) under sterile conditions. Warm up the minimal media to 37 °C by incubating in the shaker. Inspect the starter culture by swirling: it should be quite cloudy. Add the starter culture to the minimal media and grow the culture at 37° C with shaking until O.D.600 = 0.6 (this normally takes around 4.5-6 hrs). When the O.D.600 reaches 0.6, set the shaker temperature to 18° C and shake for another hour. Add 1 g glyphosate, 50 mg 3-F-L-tyrosine, 50 mg L-phenylalanine, and 50 mg L-tryptophan, then induce with 0.5 mM IPTG. After induction, incubate shaking culture at 18 °C for 18-20 hrs.
Day 2: Harvest the cell culture by centrifuging at a speed of 4,000 rpm at 4 °C. Discard the media supernatant in the empty culture flask without disturbing the pellet. Scoop out cell pellet in 3 g aliquots in a clean 50 mL falcon tube, and store in -80°C freezer indefinitely until purification.
You now have a pellet containing your protein of interest with >80% of 3-fluorotyrosine uniformly labeled throughout the sequence. Proceed to the purification protocol optimized for the unlabeled protein.
Protocol #2: expression protocol for 5-fluorotryptophan labeled protein
By Yuan Yang, edited by Robert Peterson and Mark Arbing – 08/2020 – UCLA
*This protocol has actually been performed at UCLA by Yuan Yang.
Overview: E. coli BL21 cells harboring an appropriate expression plasmid are grown in minimal media. 5-fluoroindole is added just prior to induction by IPTG. After induction the 5-fluoroindole is incorporated into the expressed protein as 5-fluorotryptophan. The protein can be purified normally.
Detailed Protocol:
Day 0: Perform a plasmid transformation into E. coli BL21-Gold (DE3) competent cells and plate on LB-Agar supplemented with antibiotic. Incubate the plate at 37 °C for 16-20 hrs. Prepare 1 L of minimal media in 2.8 L baffled culture flasks by diluting 10X M9 salts (refer to steps 1 and 2 in the minimal media recipe below) with deionized water, add 1 g of NH4Cl, autoclave with liquid cycle and store at room temperature. If you wish to generate protein with both 15N and 5-FW label, use 1g of U-15NH4Cl instead of regular NH4Cl.
Day1: Inoculate a 5 mL starter culture at 9am in LB-Antibiotics by picking up a single colony from the transformation plate or by scraping a chunk of glycerol stock. Incubate starter culture by shaking at 205 rpm in a 37 °C shaker. The duration of starter culture shaking should be >3hrs. You can also shake the starter culture overnight (~6-12 hrs).
Add the rest of reagents to the prepared 1 L minimal media (step 3 in the minimal media recipe below) under sterile conditions. Warm up the minimal media to 37 °C by incubating in the shaker. Inspect the starter culture by swirling: it should be quite cloudy. Add the starter culture to the minimal media and grow the culture at 37° C with shaking until O.D.600 = 0.6 (this normally takes around 4.5-6 hrs). When the O.D.600 reaches 0.6, set the shaker temperature to 18° C and shake for another hour. Add 60 mg of 5-fluoroindoleref (dissolved fresh in 200μL DMSO) then induce with 0.5 mM IPTG. After induction, incubate shaking culture at 18 °C for 18-20 hrs.
Day 2: Harvest the cell culture by centrifuging at a speed of 4,000 rpm at 4 °C. Discard the media supernatant in the empty culture flask without disturbing the pellet. Scoop out cell pellet in 3 g aliquots in a clean 50 mL falcon tube, and store in -80°C freezer indefinitely until purification.
You now have a pellet containing your protein of interest with >80% of 5-fluorotryptophan uniformly labeled throughout the sequence. Proceed to the purification protocol optimized for the unlabeled protein.
Protocol #3: protocol for labeling cysteine residues with fluorine
By Robert Peterson and Mark Arbing – 8/2020 – UCLA
*This protocol was adapted from the literature and has not been tried at UCLA
Overview: the cysteine residues of a purified protein are labeled with fluorine. Protein is reacted with 4,4’-dithiodipyridine (4-PDS), then with trifluoroethylthio (TET).
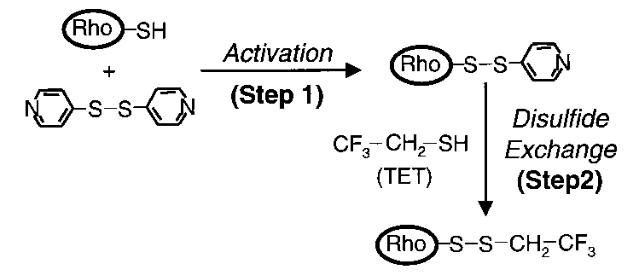
All cysteines on the protein are conjugated with TET, which contains a CF3 group.
Detailed Protocol:
Start with purified protein which is stably bound on a column. In the references, the authors were working with rhodopsin which was purified using 1D4-Sepharose beads. The entire TET labeling procedure was performed in a batch binding format with the protein bound to the Sepharose beads. One type of column that’s suited to this purpose is the Econo-Pac style columns from Bio-Rad. They are convenient to use for gravity-flow chromatography, and they contain end caps to facilitate inversion and agitation during the reactions.
Example: TET labeling 10 mg of purified rhodopsin bound to 1D4-Sepharose beads:
Suspend the beads with bound protein in 40 mL of [2 mM NaH2PO4/Na2HPO4 (pH 6), 0.05% dodecylmaltoside]. Add 4-PDS from a 1 M stock solution in ethanol to give a final concentration of 1 mM. After nutation for 5 min at room temperature, remove excess reagent by multiple washing with 40 mL of the same buffer [2 mM NaH2PO4/Na2HPO4 (pH 6), 0.05% dodecylmaltoside] under slight Argon pressure. Use a total of at least 35 times the column volume of this buffer. To be assured of complete removal of 4-PDS, the eluate can be tested spectrophotometrically by measuring the absorbance at 323 nm.
Resuspend the beads in 30 mL of 2 mM NaH2PO4/Na2HPO4 (pH 6), 0.05% dodecylmaltoside] and add 3 μL of 11.2 M TET (to give a final concentration of ~1 mM). After several hours of end-over-end agitation at room temperature, remove excess reagent as described above for 4-PDS removal. Absence of TET in the eluate can be checked spectrophotometrically by reaction with 4-PDS.
M9 Minimal media recipe
By Henry Chan – 08/23/2017 – UCLA – Feigon lab
Stock solutions:
- Thiamin, 1% sterile, filtered
- MgSO4, 1 M, sterile filtered
- FeCl3, 1M, sterile filtered
- 1000 X trace elements solution, sterile filtered
- CaCl2, 0.11%
- CoCl2, 30 uM
- CuSO4, 10 uM
- H3BO3, 100 uM
- MnSO4, 80 uM
- Na2MoO4, 3 uM
- ZnSO4, 10 uM
- NiSO4, 100 uM
- Glucose, 20% (w,v), sterile filtered (for 13C, add 2g per ½ liter of media)
- Antibiotic
Preparation:
- For 1 L of media, prepare the following in a culture flask (Use this if no 10X M9 stock):
| Reagent | 1X | 10X | |
|---|---|---|---|
| NaCl | 0.5 g | 5 g | |
| K2PO4 | 3.0 g | 30 g | |
| Na2HPO4 | 6.0 g | 60 g | |
| or | Na2HPO4 • 2H20 | 7.52 g | 75.2 g |
| or | Na2HPO4 • 7H20 | 11.32 g | 113.2 g |
| NH4Cl or 15NH4Cl | 1.0 g | 10. g |
Dilute 100mL 10X M9 with 870mL of water, and then add 1.0 g of NH4Cl or 15NH4Cl
2. Autoclave the solution
3. After autoclaving*, add the following:
| Trace elements solution | 1 mL |
| 20% Glucose | 25 mL |
| 1% Thiamin | 800μL |
| 1 M MgS04 | 2 mL |
| 1 M FeCl3 | 100μL |
| Antibiotic |
References
5-F-Trp labeling using 5-F-indole and F-labeled aromatics
- Simple and inexpensive incorporation of 19F-Tryptophan for protein NMR spectroscopy, Peter B. Crowley, Ciara Kyne and William B. Monteith, Chem. Commun., 2012, 48, 10681–10683.
F-labeled aromatics
- Aromatic 19F-13C TROSY: a background-free approach to probe biomolecular structure, function, and dynamics. Boeszoermenyi, A., et al. Nature Methods, 2019, 16, 333-340.
TET labeling of cysteines
- NMR spectroscopy in studies of light-induced structural changes in mammalian rhodopsin: Applicability of solution 19F NMR. Klein-Seetharaman, J., et al. Proc. Natl. Acad. Sci. USA, 1999, 96(24), 13744-13749.
- Solution 19F nuclear Overhauser effects in structural studies of the cytoplasmic domain of mammalian rhodopsin. Loewen, M. C., et al. Proc. Natl. Acad. Sci. USA, 2001, 98(9), 4888-4892.